Role of aaRS complexes in translation
Several studies have revealed the involvement of multi aminoacyl-tRNA synthetase complexes (MSC) in translation. In eukaryotes these complexes tend to be larger than those discovered in bacteria and archaea and also perform a wider range of functions that include both aminoacylation and noncanonical roles beyond translation.
In archaeal methanogens, seryl-tRNA synthetase:arginyl-tRNA synthetase (SerRS:ArgRS) assembly is formed to enhance aminoacylation of serine-specific tRNAs especially under conditions of elevated temperature and salt concentration. Thus, such aaRSs complexes may constitute a part of thermo- and osmoadaptation mechanisms of thermophilic methanogenic archaea, by providing an optimal microenvironment that facilitates stable tRNA aminoacylation. Since each of the aaRSs are also involved in binary complexes with their cognate tRNAs, dynamic interplay between possible functional states are currentlly investigated. The SerRSs from methanogenic archaea differ markedly from their bacterial-type counterparts. An interesting question is whether observed regulation of enzyme activity via synthetase complex formation is an exclusive ability of methanogenic-type SerRSs. If so, this feature may influence evolutionary conservation of methanogenic-type SerRS forms in archaea that live in harsh environments. Similar rationale complies with projects in which we explore possibilities of improved aminoacylation error correction by associated aaRSs vs. unassociated enzymes and investigation whether new modes of aminoacylation proofreading parallel synthetase assembling.
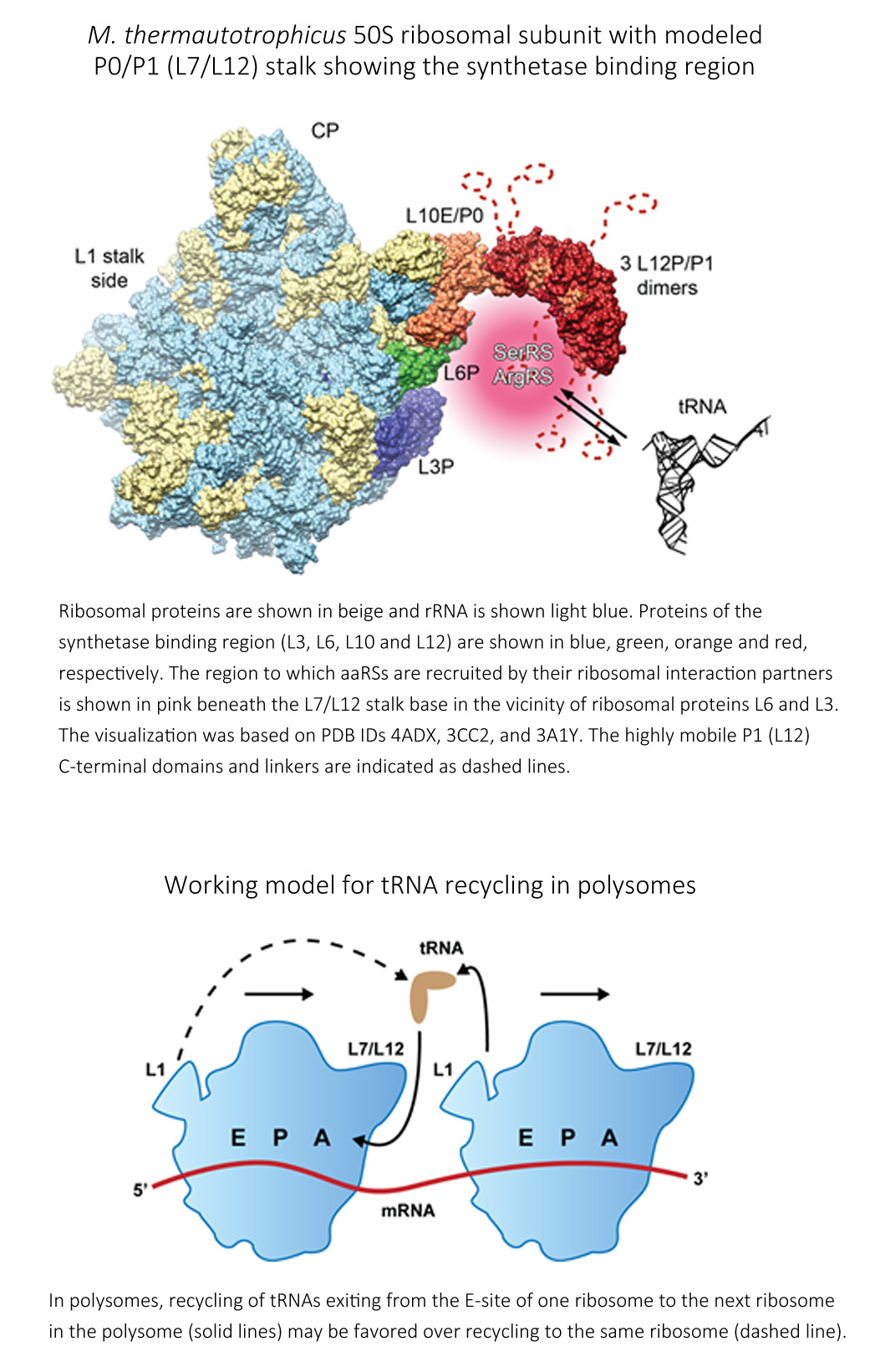
Further, atypical methanogenic-type SerRS and an archaeal ArgRS bind to the large ribosomal subunit with micromolar affinities. We are interested in detailed study of SerRS:ArgRS complex binding to Methanothermobacter thermautotrophicus ribosomes. The interpretation of data is facilitated by cryo-EM reconstructed 3D structure of M. thermautotrophicus 50S ribosomal subunit. The interaction point with ribosome was narrowed down to several ribosomal proteins. All of them comprise the P0/P1 stalk and proteins near the P0/P1 stalk base of the 50S subunit. The interior of the cell is highly crowded with macromolecules, and tRNAs may not diffuse far after exiting the ribosome. Indeed, they may remain in the vicinity of the ribosome to be recharged and reused for another cycle of elongation.
Therefore, the codon choice at the next instance of the same amino acid may be influenced by the previous codon upstream, caused by the autocorrelation of codon pairs read by the same tRNA that could be generally advantageous for the translation process. While studying coding sequences of M. thermautotrophicus genome, a bias was revealed of clustered synonymous codons, called codon co-occurrence bias. It was shown that not only the overall frequency of synonymous codons but the order in which they reside in a gene are biased. Instead of a random distribution of synonymous codons on a coding sequences, there is a bias to cluster synonymous codons for serine (Ser) or arginine (Arg) that are recognized by the same tRNA (i.e., identical codons and isoaccepting codons), indicating a possibility of tRNA re-use on the ribosome. That result supports a mechanism in which the deacylated tRNAs may be recharged by aaRSs bound to the ribosome and reused at the next occurrence of a codon encoding the same amino acid. We proposed a mechanism of tRNA recycling in which the archaeal mSerRS and ArgRS associate with P0/P1 stalk region of the ribosome to recapture the tRNAs released from the preceding ribosome in the polysomes.
We are interested in biophysical description of interaction phenomenons between aminoacyl-tRNA synthetases and other proteins by fluorescence spectroscopy, surface plasmon resonance (SPR) and Microscale Thermophoresis (MST). Generally, these results will reveal how different steps are functionally integrated to optimize ribosomal translation of the genetic code.
Selected Publications:
Archaeal aminoacyl-tRNA synthetases interact with the ribosome to recycle tRNAs.
Nucleic acids research. 42 (2014), 8; 5191-5201
Godinic-Mikulcic Vlatka; Jaric Jelena; Greber J Basil; Franke Vedran; Hodnik Vesna; Anderluh Gregor; Ban Nenad; Weygand-Durasevic Ivana.
Cryo-EM Structure of the Archaeal 50S Ribosomal Subunit in Complex with Initiation Factor 6 and Implications for Ribosome Evolution.
Journal of molecular biology. 418 (2012), 3/4; 145-160
Greber Basil J; Boehringer Daniel; Godinic-Mikulcic Vlatka; Crnkovic Ana; Ibba Michael; Weygand-Durasevic Ivana; Ban Nenad.
An Archaeal tRNA-Synthetase Complex that Enhances Aminoacylation under Extreme Conditions.
The Journal of biological chemistry. 286 (2011), 5; 3396-3404
Godinic-Mikulcic Vlatka; Jaric Jelena; Hausmann Corinne D; Ibba Michael; Weygand-Durasevic Ivana.