Plant aminoacyl-tRNA synthetases
Aminoacyl-tRNA synthetases (aaRSs) play a crucial role in translation by catalyzing the ligation of their cognate amino acids and tRNAs to establish the genetic code. Beyond their central role in translation, aaRSs have evolved a wide array of alternative functions both inside and outside translation. These noncanonical functions are often accomplished by interaction of aaRSs with other proteins. Plant aaRSs are less studied than aaRSs from other organisms. There is only one report on alternative function of plant aaRS outside translation. In plants, aaRSs are required for translation in three different compartments of the plant cell: chloroplasts, mitochondria and the cytosol.
Our work focuses on plant aminoacyl-tRNA synthetases with special emphasis on their substrate recognition and specificity, fidelity, cellular localization, evolution, interactions with other proteins, as well as their role in abiotic stress response. We are particularly interested in seryl-tRNA synthetases (SerRS) from Arabidopsis thaliana, model plant organism and maize, agronomically important plant species.
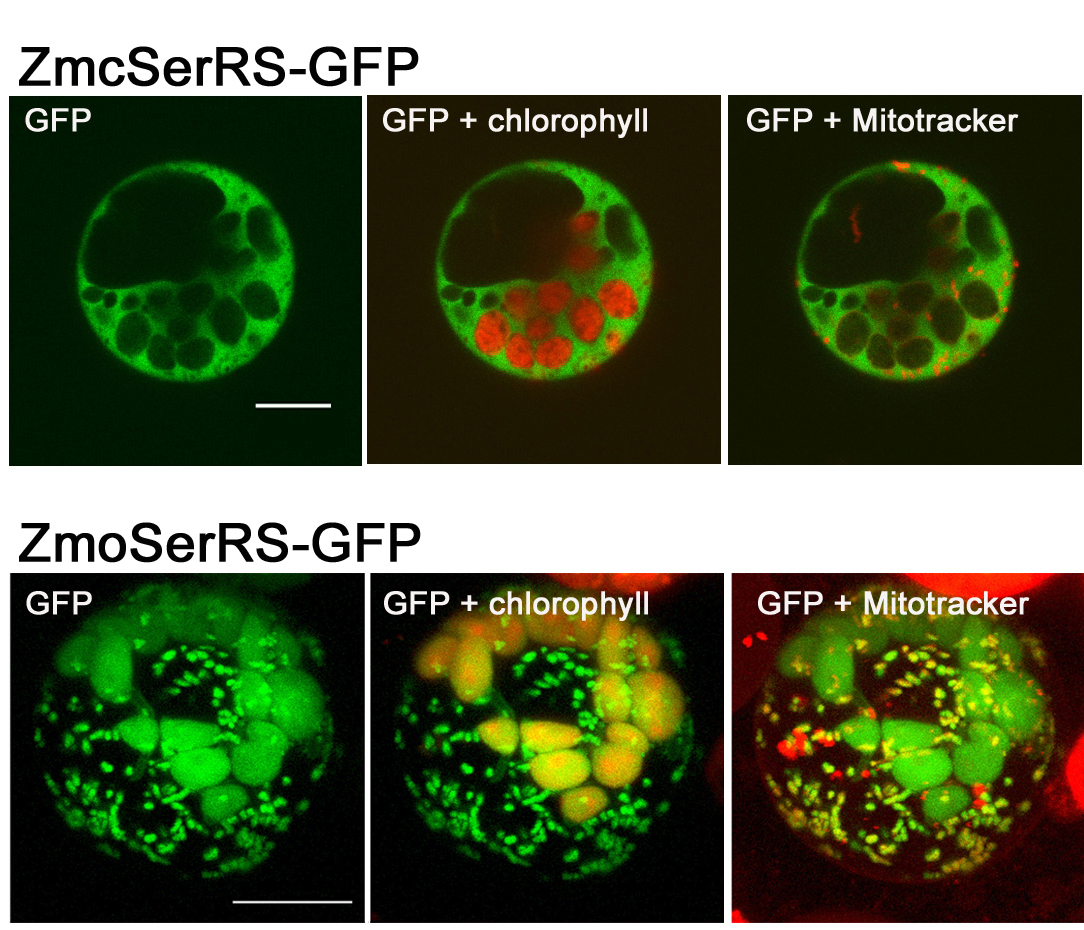
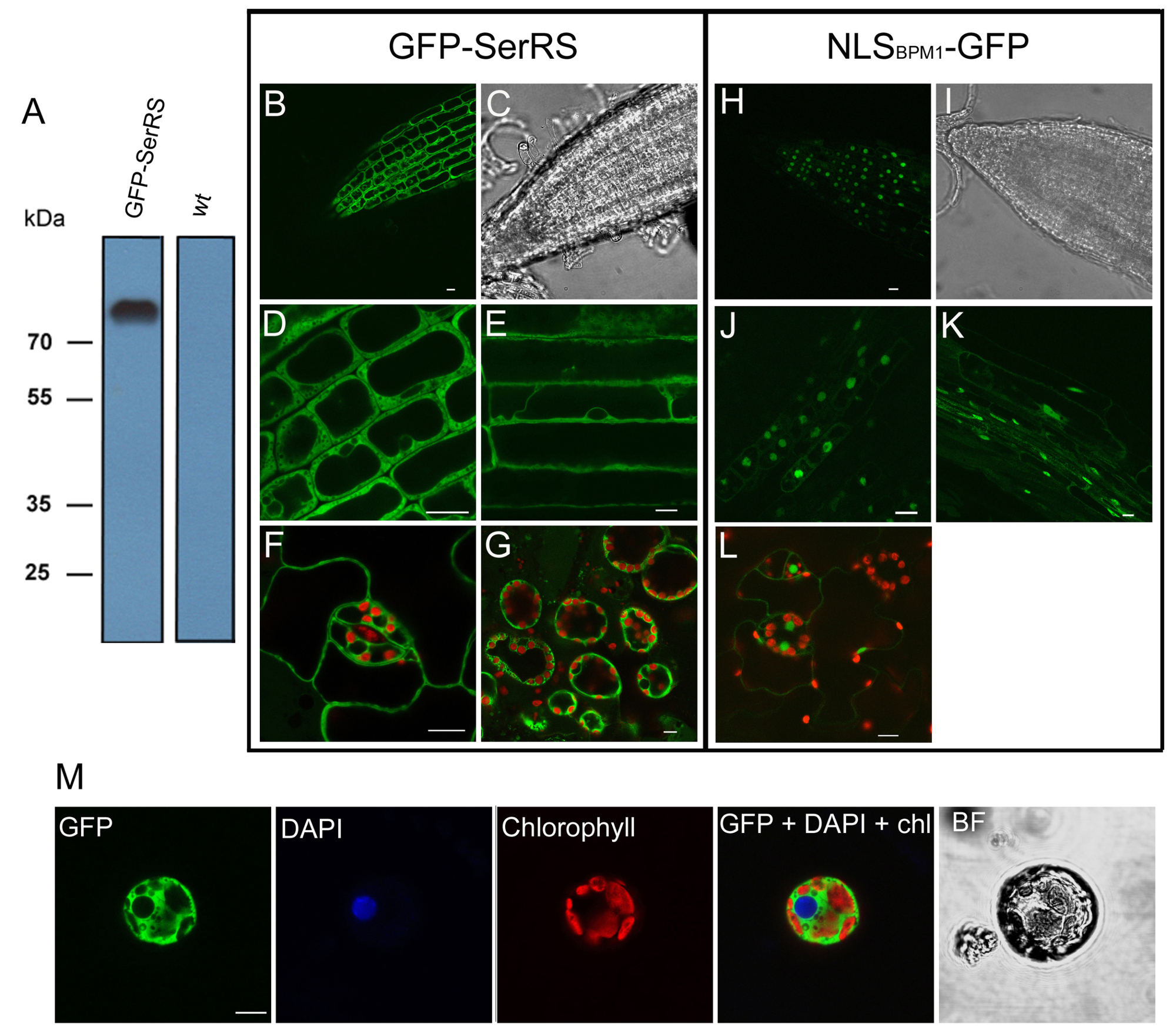
In plants there are two SerRS proteins. We showed that one of them is dually targeted to chloroplasts and mitochondria, while the other is exclusively located in cytosol. Cytosolic SerRS of eukaryotic origin shows exceptionally broad tRNASer specificity and flexibility, recognizing both eukaryotic and bacterial tRNAsSer that display different secondary and tertiary structures. Both cytosolic and organellar SerRS enzymes display high fidelity in amino acid recognition which is established by highly selective synthetic pathways in corroboration with hydrolytic proofreading.
Recently, we identified several potential protein-interacting partners of plant SerRS using two highthroughput interactome technologies, TAP-MS and Y2H. In addition, transgenic Arabidopsis seedlings overexpressing SerRS grow better than wild type plants under various conditions of abiotic stress. These observations indicate noncanonical role of SerRS.
Our current research is directed towards:
- characterization of SerRS protein interactors in plants
- investigation of SerRS role in plant stress response
Localization of two maize seryl-tRNA synthetases ZmcSerRS and ZmoSerRS visualized by confocal fluorescent microscopy. Proteins fused with GFP were transiently expressed in maize protoplasts. Upper panel: ZmcSerRS is localized in cytosol. Lower panel: Dual targeting of organellar ZmoSerRS to chloroplasts and mitochondria.
Exclusive cytosolic localization of Arabidopsis SerRS in transgenic Arabidopsis seedlings overexpressing GFP-SerRS. As a control for nuclear localization, Arabidopsis seedlings overexpressing nuclear localization signal of BPM1 protein fused to GFP (NLSBPM1-GFP) are analyzed. (A) Detection of the intact GFP-SerRS fusion protein (75 kD) in total protein extracts from transgenic Arabidopsis seedlings by western blot analysis. Total protein extract from wild type plants (wt) was used as negative control. (B-G) Transgenic seedlings expressing GFP-SerRS. (H-L) Transgenic seedlings expressing NLSBPM1-GFP. (B, C, H, I) Root tip. (D, J) Enlarged view of root elongation zone. (E) Root epidermal cells. (F, L) Leaf epidermis with stomata. (G) Leaf mesophyll cells. (K) Longitudinal view of the root. (M) Protoplast isolated from transgenic seedling expressing GFPSerRS.
Selected Publications:
Exclusive cytosolic localization and broad tRNASer specificity of Arabidopsis thaliana seryl-tRNA synthetase.
Journal of Plant Biology. 59 (2016), 1; 44-54
Kekez Mario; Bauer Natasa; Saric Ela; Rokov-Plavec Jasmina.
Substrate recognition and fidelity of maize seryl-tRNA synthetases.
Archives of Biochemistry and Biophysics. 529 (2013), 2; 122-130
Rokov-Plavec Jasmina; Lesjak Sonja; Gruic-Sovulj, Ita; Mocibob Marko; Dulic Morana; Weygand-Durasevic, Ivana.
Dual targeting of organellar seryl-tRNA synthetase to maize mitochondria and chloroplasts.
Plant Cell Reports. 27 (2008), 7; 1157-1168
Rokov-Plavec Jasmina; Dulic Morana; Duchêne Anne-Marie; Weygand-Durasevic Ivana.